The qualitative simulation method has been implemented in Java in a computer tool called Genetic Network Analyzer (GNA). GNA has been used for the modeling and simulation of several well-studied regulatory processes, such as the initiation of sporulation in Bacillus subtilis. By using the method, we have been able to reproduce the essential characteristics of the choice between vegetative growth and sporulation, in particular the role played by positive and negative feedback loops. We have also started the application of the method to genetic regulatory systems of which the functioning is less understood, such as the nutritional stress response in Escherichia coli and Synechocystis PCC 6803.
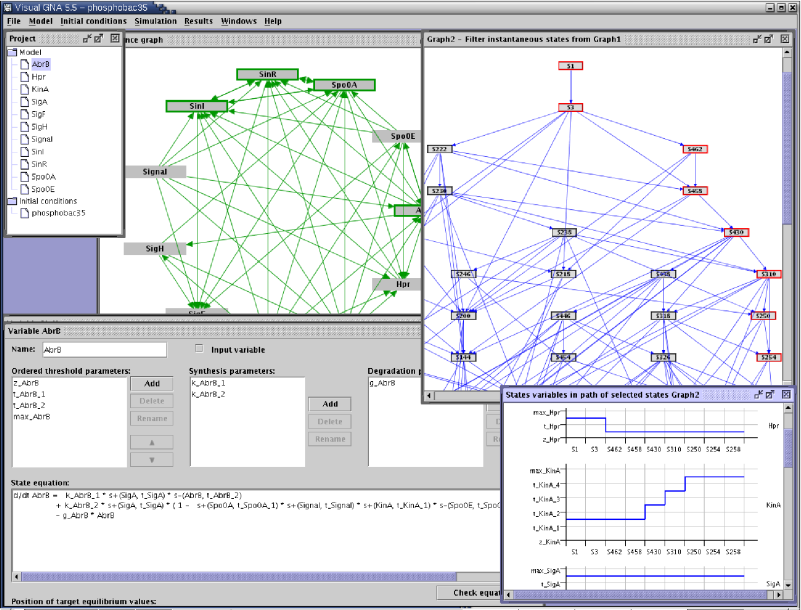
Graphical interface of the qualitative simulation tool Genetic Network Analyzer (GNA). |
Literature
H. de Jong, J. Geiselmann, G. Batt, C. Hernandez, M. Page (2004), Qualitative simulation of the initiation of sporulation in Bacillus subtilis, Bulletin of Mathematical Biology, 66(2):261-300.
H. de Jong, J. Geiselmann, C. Hernandez, M. Page (2003), Genetic Network Analyzer: Qualitative simulation of genetic regulatory networks, Bioinformatics, 19(3):336-344.
H. de Jong, J. Geiselmann, D. Thieffry (2003), Qualitative modelling and simulation of developmental regulatory networks, S. Kumar, P.J. Bentley (eds), On Growth, Form, and Computers, Academic Press, London, 109-134. |