The main key word for describing GenoStar is integration : integration of data, of methods for analysing these data and of modules structuring these methods into strategies.
Data integration
GenoStar is built on top of GenoCore which provides advanced services for data modeling and management. All the data which are handled by the methods within GenoStar are explicitely described in an entity-relationship data model (AROM). Type and integrity checking is extensive : every method "knows" the type of data it expects as input and the type of results it computes. Moreover, these UML-like descriptions play an important role for documenting the modules ; their extension, through the addition of new methods, is thus made easier and more secure.
Method integration
A frequently asked question to the designers of GenoStar is : Why do you bother in developping such a platform when fast and efficient bioinformatic methods are freely available on Internet ? The answer is that integrating methods within a plateform grealy increases the ease with which data can be passed on between methods. Since all the data are explicitely and formally described, the problem of format transformation disappears. Moreover, mismatches between data types may be solve automatically.
Just remember how much of their time biologists and bioinformaticians do spend in writing scripts to make their data suitable to the available methods. Since complex analyses imply more than one method, GenoStar provides pre-defined strategies which organise methods into pipe-lines. The next versions of GenoStar will allow the users to define new strategies, or to modify existing ones, in a very simple way, relying on a powerful graphical programming interface. Another interface will display the state of computations and the flow of data, so that the user will be able to follow the execution process and act on it if required.
Module integration
The present version of GenoStar includes three modules, but the software architecture has been designed in such a way that the addition of new modules is straightforward. A practical example was provided when GEB (GenoExpertBacteria), intially designed as a stand-alone module, has been integrated in GenoStar in a few working days. The next version of GenoStar (1.3), will allow its users to quickly define, developp and integrate new modules dedicated to their specific problematics.
The modules can share data and results. As a concrete example, which is detailed in the tutorial, data on genes can be exchanged between GenoAnnot, where they have been identified, to GenoBool, where the application of classification methods over the sequences reveal clusters of genes, and sent back to GenoAnnot where the genomic location of the genes of the clusters give a decisive clue to the interpretation of these clusters and set an interesting biological question.
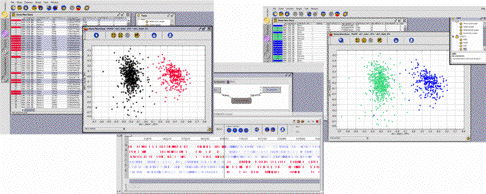 On the basis of the way their sequences make use of the genetic code, the set of genes of the bacterium B. burgdorferi has been partitioned into two clusters with the help of a classification method of the GenoBool module (left screen shot). Coming back to the GenoAnnot module, which produced the prediction of the gene locations and therefore their sequences, the user will see (centre) that the genes of a same cluster tend to be located either on the leading or on the lagging strand. He may then formulate an hypothesis, validate it through an explicit request on the set of genes (right screen shot), and search for a biological explanation to this correlation between a physico-chemical property and the codon usage (this correlation has been discovered in 1998 by James O. McInerney: "Replicational and transcriptional selection on codon usage in Borrelia burgdorferi" PNAS 1998; 95: 10698-10703). | |