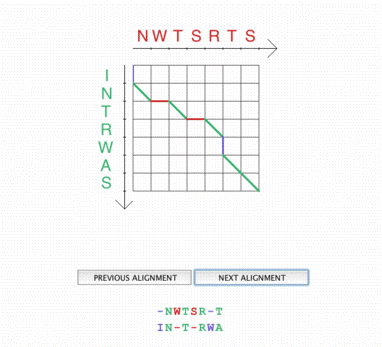
 A path in the score matrix and the associated aligment A path in the score matrix and the associated aligment |
The Needleman - Wunsch dynamic programming algorithm module offers different vizualisation levels. On the first one, the user visualizes the path in the distance matrix associated with each possible alignment, as a result of the execution of the algorithm on two given sequences. The distance matrix can be displayed and filled step by step. Each computation step is graphically explained.
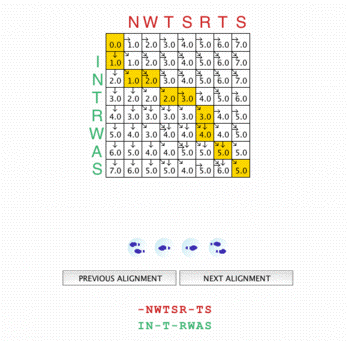 The score matrix The score matrix |
Once the matrix has been completely filled, the pathway is constructed, to give the best scoring alignment. The student is also asked to construct himself a path that leads to an optimal alignment. |