The HERBS current user interface is composed of:
1. A main window
 inviting the user to choose metabolic pathway(s) and organism(s) to query (cf figure 1). inviting the user to choose metabolic pathway(s) and organism(s) to query (cf figure 1). 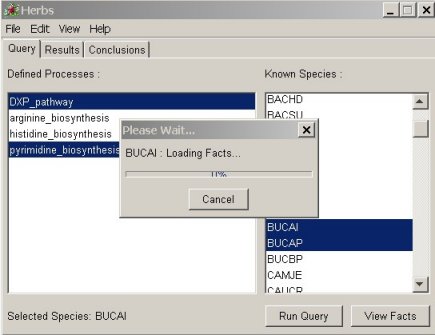 figure 1: figure 1:The user has the hability to select local or up to date data from a specific database. |
 showing the results into the form of visualization trees (cf figure 2). showing the results into the form of visualization trees (cf figure 2).
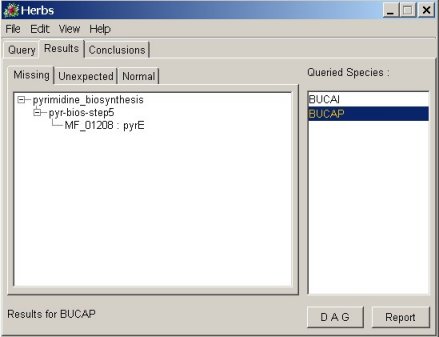 figure 2: figure 2:For each process under study, missing, unexpected and normal proteins are visible into the form of tree. |
2. A report dialog allowing the user to save results in text format(cf figure 3).  figure 3: figure 3:The results for each queried species and pathway are reported in a text file. |
3. A graphical window displaying the results into DAG (Directed Acyclic Graph) view (cf figures 4 & 5).
An example of the detection of a missing protein is illustrated by the following results. The root of the DAG corresponds to the biological process under study, the internal nodes correspond to the steps of this process and the other ones to the proteins which are implied at each step. An adapted colour code indicates the inconsistencies located by the HERBS engine: any orange part of the DAG indicates a problem.
The presence of a specific metabolic pathway has been tested in two bacteria:
 BUCAI: Buchnera aphidicola (subsp. Acyrthosiphon pisum) (Acyrthosiphon pisum symbiotic bacterium). BUCAI: Buchnera aphidicola (subsp. Acyrthosiphon pisum) (Acyrthosiphon pisum symbiotic bacterium).
 BUCAP: Buchnera aphidicola (subsp. Schizaphis graminum)*. BUCAP: Buchnera aphidicola (subsp. Schizaphis graminum)*.
The two microbial organisms are assumed to be really closely related, especially in terms of metabolic processes. One thus expects to find all proteins implied in the pathway under study, pyrimidine boisynthesis (cf rules defining this pathway in Herbs engine), in both organisms.
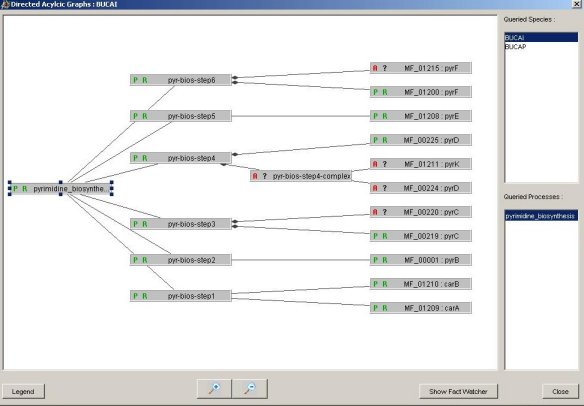 figure 4: figure 4:The HERBS representation of the pyrimidine biosynthesis in BUCAI. All expected proteins are present and thus appear in gray. |
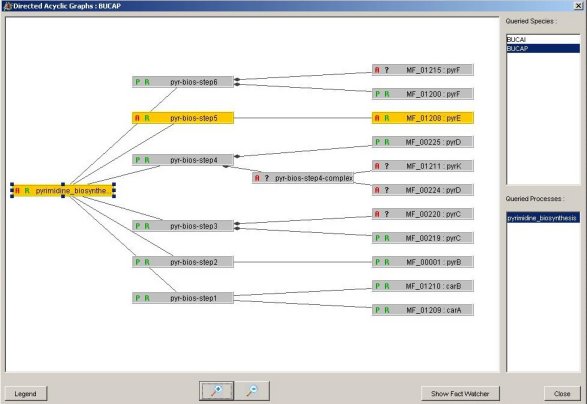 figure 5: figure 5:The HERBS epresentation of the pyrimidine biosynthesis in BUCAP. In this case, it is easy to notice the inconsistencies which appear in orange. The pyrE expected protein is actually absent, subsequently the step 5 of the pathway cannot append. |
At this stage of BUCAP proteome analysis, several hypothesis can be expressed:
 The pyrE protein is absent because of prediction errors of the CDS (Coding Sequence) for this protein (sequencing problem, frameshifts...). The pyrE protein is absent because of prediction errors of the CDS (Coding Sequence) for this protein (sequencing problem, frameshifts...).
 BUCAP uses an alternative process to achieve the pyrimide biosynthesis. BUCAP uses an alternative process to achieve the pyrimide biosynthesis.
 other ideas? other ideas?
The biologist is now warned up of the inconsistencies in the BUCAP proteome. He can continue this analysis by using other tools such as GEB to explore the corresponding nucleic sequence.
*More information of BUCAI and BUCAP organisms can be found on HAMAP site. |