The PSTs, which have been automatically generated from MS/MS spectra analysis, are submitted to a second software component: PepMap. PepMap is responsible of mapping the PSTs with the contents of protein databases (or, directly, to complete translated chromosomes).
PepMap algorithm consists in two steps:
 Mapping a PST on a polypeptide sequence consists in searching for the PST sequence part, then checking that at least one of the flanking masses corresponds to the adjacent sequences. Mapping a PST on a polypeptide sequence consists in searching for the PST sequence part, then checking that at least one of the flanking masses corresponds to the adjacent sequences.
 Mapping of a PST on a protein sequence Mapping of a PST on a protein sequence |
 Protein identification results in extracting proteins on which the more PST matches occur. Protein identification results in extracting proteins on which the more PST matches occur.
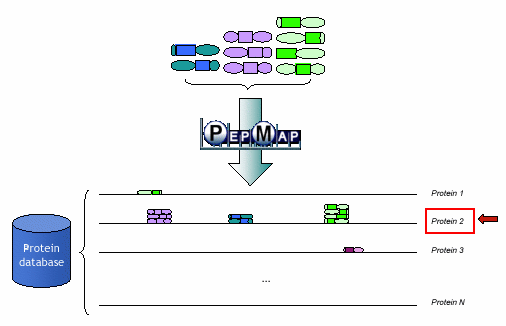 PepMap protein identification overview PepMap protein identification overview | |