A variety of methods for the modeling and simulation of genetic regulatory networks have been proposed, such as approaches based on differential equation models and stochastic models. These models provide detailed descriptions of genetic regulatory networks, down to the molecular level. In addition, they can be used to make precise, numerical predictions of the behavior of regulatory systems. Many excellent examples of the application of these methods to prokaryote and eukaryote networks can be found in the literature.
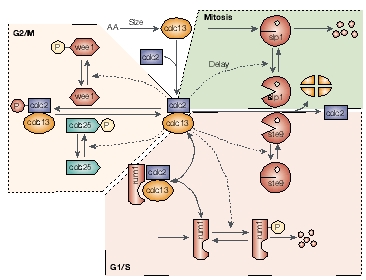
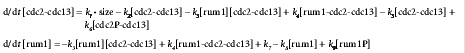
Cell-cycle control system in fission yeast and equations describing the dynamics of the proteins cdc2-cdc13 and rum1.J.J. Tyson et al. (2001), Nat. Rev. Mol. Cell. Biol., 2:908-916. |
In many situations of biological interest, however, the application of differential equation and stochastic models is seriously hampered. In the first place, the biochemical reaction mechanisms underlying regulatory interactions are usually not or incompletely known. In the second place, quantitative information on kinetic parameters and molecular concentrations is only seldom available, even in the case of well-studied model systems.
Literature
H. de Jong (2002), Modeling and simulation of genetic regulatory systems: A literature review, Journal of Computational Biology, 9(1):69-105.
J. Hasty, D. McMillen, F. Isaacs, J.J. Collins (2001), Computational studies of gene regulatory networks: in numero molecular biology, Nature Review Genetics, 2(4):268-279.
P. Smolen, D.A. Baxter, J.H. Byrne (2000), Modeling transcriptional control in gene networks: Methods, recent results, and future directions, Bulletin of Mathematical Biology, 62(2):247-292.
H. de Jong, D. Ropers, C. Chaouiya, D. Thieffry (2005), Modélisation, analyse et simulation de réseaux de régulation génique, Biofutur, 252:36-40. |