Drawing on an explicit description of biological entities and their structures, GenoAnnot is able to guide the user through the choices and combinations of methods of annotation using advanced bioinformatics.
The assortment of available methods is large and easily extendable. They include lexical or structural pattern searching, Markov chains and databank screening. A dedicated interface allows the user to view the results of the analysis, to append comments and to validate them. The annotation process of a sequence can be memorized and can be played back after modifying the parameters of a particular method.
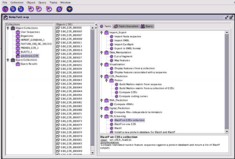
Strategies are pre-defined combinations of methods. Genostar offers a wide range of preset strategies, enabling complex analytical tasks to be carried out under the complete control of the user. New strategies can be easily added, for example by making simple changes to existing ones.
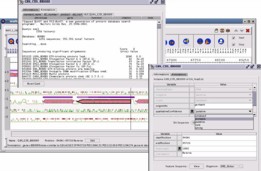
Results produced by analysis of a genomic sequence are visually synthesized on GenoStar cartographic interface. The user can view predictions originating from different methods to help with the final selection. Every graphic object has a knowledge base object associated with it, and related information can be captured.
GenoAnnot as available in the current version of GenoStar can carry out the annotation of a complete bacterial genome. |