The qualitative simulation method is currently being applied by D. Ropers to genetic regulatory networks of which the functioning is only partially understood. We collaborate with the laboratories of J. Geiselmann (université Joseph Fourier) and J. Houmard (ENS Paris) on the modeling of the global regulation of transcription in the bacteria Escherichia coli and Synechocystis PCC 6803. We try to understand how the response of the bacterium to a nutritional stress emerges from the network of interactions between its genes, proteins, and small molecules. In order to elucidate this network, we use the computer tool Genetic Network Analyzer (GNA) in combination with experimental methods for testing the predictions of the simulator, as well as classical bioinformatics approaches, such as sequence analysis. This work is supported by the European project Hygeia: Hybrid Systems for Biochemical Network Modeling and Analysis (NEST-2003-1 ADVENTURE).
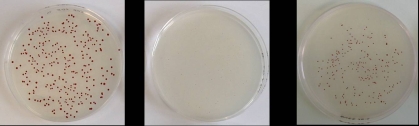
Three E. coli cultures grown overnight: wild-type (left), crp mutant (middle), and crp and suppressor mutant (right). The gene crp encodes the protein CRP, a global regulator of transcription.Pictures kindly provided by J. Geiselmann |
Literature
H. de Jong, J. Geiselmann, G. Batt, C. Hernandez, M. Page (2003), Qualitative simulation of the initiation of sporulation in Bacillus subtilis, Bulletin of Mathematical Biology, 66(2):261-300.
D. Ropers, H. de Jong, M. Page, D. Schneider, J. Geiselmann (2004), Qualitative simulation of the nutritional stress response in Escherichia coli, INRIA, RR-5412. |