It is now commonly accepted that most interesting properties of an organism emerge from the interactions between its genes, proteins,metabolites, and other constituents. This implies that, in order to understand the functioning of an organism, we need to elucidate the networks of interactions involved in gene regulation, metabolism,signal transduction, and other cellular and intercellular processes.
Genetic regulatory networks control the spatiotemporal expression of genes in an organism, and thus underlie complex processes like cell differentiation and development in prokaryotic and eukaryotic organisms. Genetic regulatory networks consist of genes, proteins,metabolites, and other small molecules, as well as their mutual interactions. Their study has taken a qualitative leap through the use of modern genomic techniques that allow simultaneous measurement of the expression of all genes of an organism.
In addition to experimental tools, mathematical methods supported by computer tools are indispensable for the analysis of genetic regulatory networks. As most networks of interest involve many genes connected through interlocking positive and negative feedback loops, an intuitive understanding of their dynamics is difficult to obtain and may lead to erroneous conclusions. Modeling and simulation tools allow the behavior of large and complex systems to be predicted in a systematic way.
Literature
J.M. Bower, H. Bolouri (eds) (2001), Computational Modeling of Genetic and Biochemical Networks, MIT Press, Cambridge, MA.
C.P. Fall, E.S. Marland, J.M. Wagner, J.J. Tyson (eds) (2002), Computational Cell Biology, Springer-Verlag, New York.
H. Kitano (ed.) (2001), Foundations of Systems Biology, MIT Press, Cambridge, MA.
F. Rechenmann, H. de Jong (2005), Le vivant en équations, La Recherche, 383:32-37.
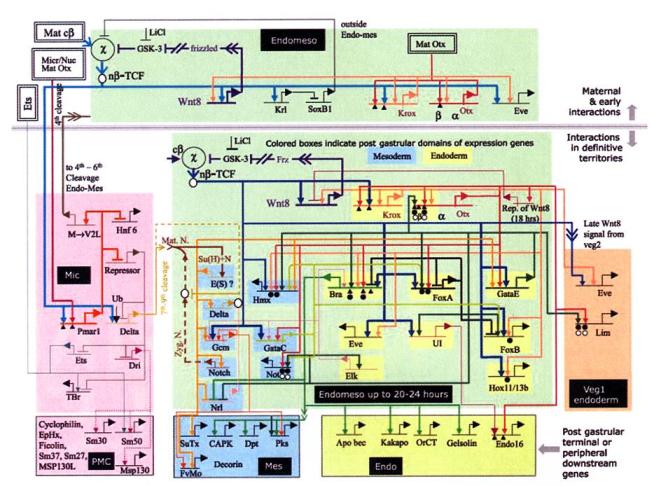 Genetic regulatory network controlling the development of the body plan of the sea urchin embryo Genetic regulatory network controlling the development of the body plan of the sea urchin embryoDavidson et al., Science, 295(5560):1669-1678. | |