A prerequisite for the simulation of genetic regulatory networks is the availability of a model of the system. Traditionally, models are hand-crafted from information in the experimental literature. However, for many systems at the forefront of biological research the information required for this is simply not available. Recent breakthroughs in the mass production of experimental data have made it possible to obtain models of genetic regulatory networks in a different way, namely by identifying the networks from time-series measurements of gene expression. In the context of the thesis of Samuel Drulhe, co-supervised by Giancarlo Ferrari-Trecate (INRIA Rocquencourt), we explore the application of methods for the identification of hybrid systems to the network inference problem. These methods are well-adapted to the piecewise-linear differential equation models underlying the qualitative simulation method. Using hybrid-systems identification methods, we will try to infer the network controlling the nutritional stress response of Escherichia coli from time-series measurements of gene expression.
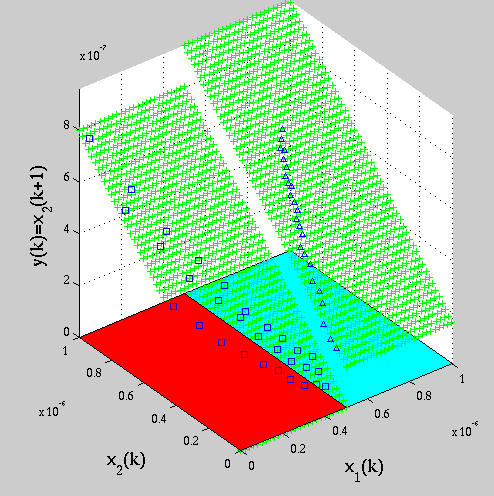 Identification results for a two-gene network: classified datapoints and estimated models in two regions of the phase space. Identification results for a two-gene network: classified datapoints and estimated models in two regions of the phase space. |
P. D'haeseleer, S. Liang, R. Somogyi (2000), Genetic network inference: From co-expression clustering to reverse engineering, Bioinformatics, 16(8):707-726.
T.S. Gardner, D. di Bernardo, D. Lorenz, J.J. Collins (2003), Inferring genetic networks and identifying compound mode of action via expression profiling, Science, 301(5629):102-105.
G. Ferrari-Trecate, M. Muselli, D. Liberati, M. Morari (2003), A clustering technique for the identification of piecewise affine systems, Automatica, 39(1):205-218. |