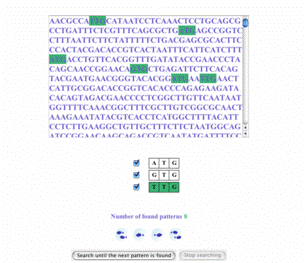
 Simple pattern matching: the search for STOP codons Simple pattern matching: the search for STOP codons |
Simple patterns, such as Start or Stop codons, are easily expressed as simple strings to be searched for in the sequence. Errors in matching may be allowed. The user can follow the sequential search process, step by step, or by skipping directly to the next occurence.
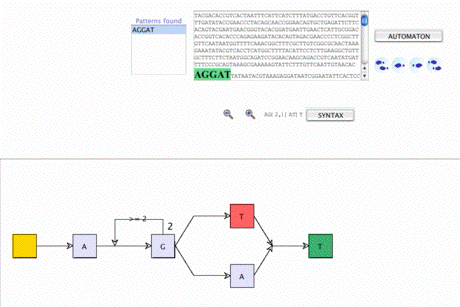 Searching CDS patterns using regular expressions Searching CDS patterns using regular expressions |
More complex patterns, such as RBS, may be expressed as regular expressions. The corresponding automaton is constructed step by step. Then, the search for a pattern in the DNA sequence matching the regular expression is made. A sub-sequence matches the regular expression if a complete path in the corresponding automaton is found. Every path in the automaton is evaluated until the last state is reached. |