The Taggor module is part of the PepLine software pipeline for proteomic analysis. It has been developed in the context of the Helix CEA/LCP collaboration.
Taggor aims at determining a set of PST candidates from a processed MS/MS spectrum (i.e. a peak-list) without complete peptide reconstruction or protein database query.
The Taggor algorithm involves three steps:
 All sequences of N amino acids (N equals 3 to 5) are generated. All sequences of N amino acids (N equals 3 to 5) are generated.
 Each of these sequences is aligned to the MS/MS spectrum according to the Y ions reading orientation. If a sequence matches a set of peaks, then flanking masses are computed and a new PST is generated. A score is defined for each PST: it consists in the product of the relative intensities of spectrum peaks that correspond to the PST anchoring sequence. Each of these sequences is aligned to the MS/MS spectrum according to the Y ions reading orientation. If a sequence matches a set of peaks, then flanking masses are computed and a new PST is generated. A score is defined for each PST: it consists in the product of the relative intensities of spectrum peaks that correspond to the PST anchoring sequence.
 Finally, a set of best PST candidates (usually 20 PSTs) is retained for further analysis. Finally, a set of best PST candidates (usually 20 PSTs) is retained for further analysis.
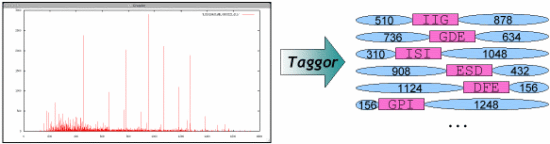 Taggor software overview Taggor software overview | |